hi-c seq dna
Hi-C results reveal chromosomal interactions such as. RNA-Sequencing high-throughput mRNA-Seq total RNA-seq 3-Tag-Seq miRNA-seq.

Hi C 3 0 Improved Protocol For Genome Wide Chromosome Conformation Capture Lafontaine 2021 Current Protocols Wiley Online Library
Hi-C is a technology to analyze the spatial structure of chromatin in cells quantifying the number of interactions between genomic loci that are adjacent in 3D space but may be separated by many nucleotides in the linear genome in this paper chromatin interactions may be just the product.
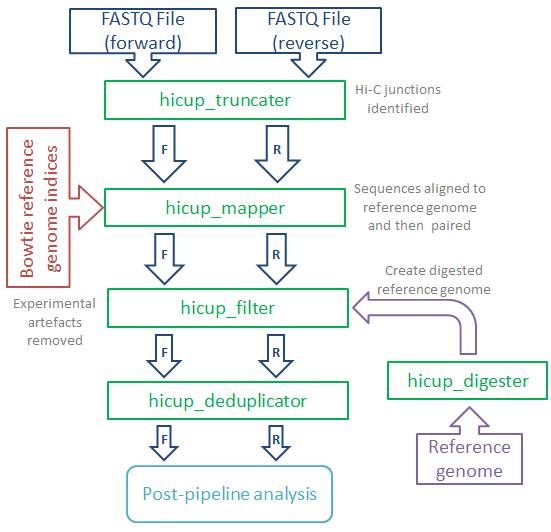
. Use 1200 Units T4 DNA Ligase to ligate the adapters to the DNA. Below we outline the principles and details of the general Hi-C protocol. In Hi-C a biotin-labeled nucleotide is incorporated at the ligation junction making it possible to enrich for chimeric DNA ligation junctions when modifying the DNA molecules for deep sequencing.
Hi-C sequencing is highthroughput chromosome conformation capture technique to analyze spatial genome organization and map higherorder chromosome folding and topological associated domains. Hi-C protocol begins by using formaldehyde to crosslink the cells which results in the covalent linking of the chromosomal loci through their protein-DNA interactions. Remove non-ligated Paired End adapters by reclaiming the Hi-C DNA bound beads and washing the beads twice with 400 μl 1x TB.
Hi-C is a popular technique to map three-dimensional chromosome conformation. Here we describe an updated protocol for Hi-C Hi-C 20 that integrates. Hi-C 可以与 RNA-SeqChIP-Seq 等数据进行联合分析从基因调控网络和表观遗传网络来阐述生物体性状形成的相关机制参考文章Hi-C 技术 Hi-C技术到底能做什么 Q3.
Chromatin conformation capture assays that are used to detect genome-wide DNADNA interactions. Hi-C Sequencing The Applied Bioinformatics Laboratories offer analysis of Hi-C sequencing or Hi-C-seq data. Hi-C 3C-Seq and Capture-C comprise a family of methods for analyzing chromatin interactions.
To test whether increased chromatin accessibility is associated with chromosome conformation variation we performed high-throughput chromosome conformation capture Hi-C sequencing in representative mutants that display different levels of DNA methylation loss including cmt2 cmt3 and ddcc and combined this analysis with previously published Hi-C data. Hi-C assay is an extension of chromosome conformation capture 3C assay studying chromosomal interactions. In Hi-C a biotin-labeled nucleotide is incorporated at the ligation junction enabling selective purification of chimeric DNA ligation junctions followed by deep sequencing.
Request a Hi-C-Seq Service To request this service from us please provide the following. Cells are fixed with formaldehyde or another fixative crosslinking DNA-DNA and DNA-protein interactions. As such for a given ligated fragment the two sequences obtained should represent two different restriction fragments that were ligated together in the.
To study the chronological order of chromatin organization and DNA methylation we analyze chromatin interactions in situ Hi-C and DNA methylation WGBS together with gene expression RNA-seq. In principle Hi-Cs resolution is only limited by the size of restriction fragments. Add 6 picomoles of Illumina Paired End adapters per μg of Hi-C DNA available for ligation.
The compatibility of Hi-C with next generation sequencing platforms makes it possible to detect chromatin interactions on an unprecedented scale. Capture-C adds an additional pull-down of the biotinylated fragments with magnetic beads to the 3C method. C DNA methylation arrays and bisulfite sequencing are used to measure DNA methylation levels.
A new refinement of the Capture-C method NG Capture-C is available. ChIP-seq to study the interaction between proteins and DNA in nuclei. Find the Right Kit.
PacBio Sequel II Library Prep. Gene Expression Profiling with 3-Tag-Seq. We present the Hi-C Interaction Frequency.
De novo genome assemblies. Next DNA is cleaved by restriction digestion and DNA ends are filled in with biotinylated. The general workflow of Hi-C and Micro-C are similar and consist of the following steps.
Thus Hi-C is the method of choice for the chromosome-scale scaffolding of genome assemblies. While 3C and its subsequent adaptations require the choice of a set of target loci Hi-C employs high-throughput sequencing and can identify genome-wide unbiased long-range interactions. Scaffolding of long read assemblies using long range contact information by Ghurye et al.
However insufficient sequencing depth forces researchers to artificially reduce the resolution of Hi-C matrices at a loss of biological interpretability. High Molecular Weight DNA Isolation HMW-DNA Hi-C Library Preparations and. Covalent crosslinking of physically proximal genomic loci Chromatin fragmentation Ligation and deep sequencing to identify interacting genomic loci The researchers found Micro-C to have higher resolution and signal-to-noise ratio than Hi-C.
Please see for example this paper. The cross-linked chromatin segment is then cut out with a restriction enzyme and the segment restriction ends are marked by filing in with biotin-labeled nucleotides 25 30. Hi-C uses high-throughput sequencing to find the nucleotide sequence of fragments and uses paired end sequencing which retrieves a short sequence from each end of each ligated fragment.
ChIP-seq CUT RUN and CUT TAG are used to identify regulatory elements using histone mark and TF enriched regions. These methods convert chromatin interactions reflecting topological chromatin structures into digital information counts of pair-wise interactions. COVIDSeq Assay 96 samples A high-performing fast and integrated workflow for sensitive applications such as human whole-genome sequencing.
Hi-C is an attractive option for querying genome-wide chromatin interactions as well as the differences found in response to signaling differentiation or disease. Nanopore Sequencing PromethION Linked Read Sequencing 10X Genomics Chromium Technology. Hi-C quantifies the number of interactions between genomic loci that are closely located in 3-D space but may be separated by many.
Chromosome conformation capture-based methods such as Hi-C have become mainstream techniques for the study of the 3D organization of genomes. The high-resolution genome-wide map of interacting genetic loci that is generated from Hi-C data can then be used across multiple genomic. 3C 4C 5C Capture-C Hi-C DNase Hi-C ChIA-PET and HiChIP are used to map chromatin interactions.
Arima Hi-C is a proximity ligation method that captures the organizational structure of chromatin in three dimensions where genomic sequences that are distal to each other in linear distance can be closer to each other in the 3D space. The two-dimensional genome organization of scaffolds can be derived from the Hi-C 3D chromatin contact data. The Hi-C approach extends 3C-Seq to map chromatin contacts genome-wide and it has also.
The compatibility of Hi-C with next generation sequencing platforms makes it possible to detect chromatin interactions on an unprecedented scale. Incubate for 2 hours at room temperature. In HIC methods cells are crosslinked with formaldehyde to link DNA regions covalently that are in close spatial proximity in nucleus.

Juicer Provides A One Click System For Analyzing Loop Resolution Hi C Experiments Cell Systems

De Novo Prediction Of Human Chromosome Structures Epigenetic Marking Patterns Encode Genome Architecture Pnas

Hi C Overview Cells Are Cross Linked With Formaldehyde Resulting In Download Scientific Diagram
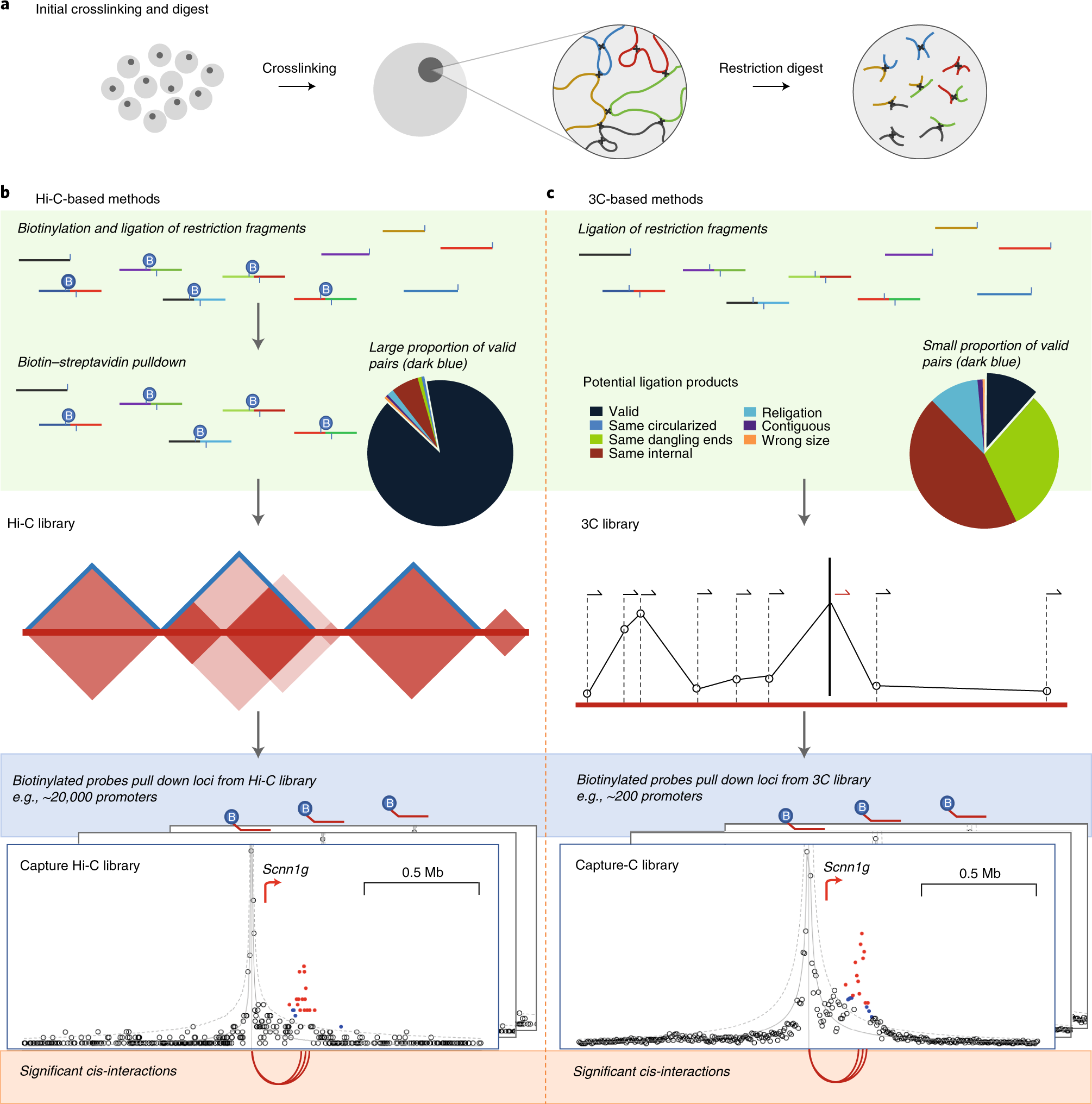
Detecting Chromosomal Interactions In Capture Hi C Data With Chicago And Companion Tools Nature Protocols

Application Of Hi C And Other Omics Data Analysis In Human Cancer And Cell Differentiation Research Sciencedirect

Babraham Bioinformatics Hicup Hi C Analysis Pipeline
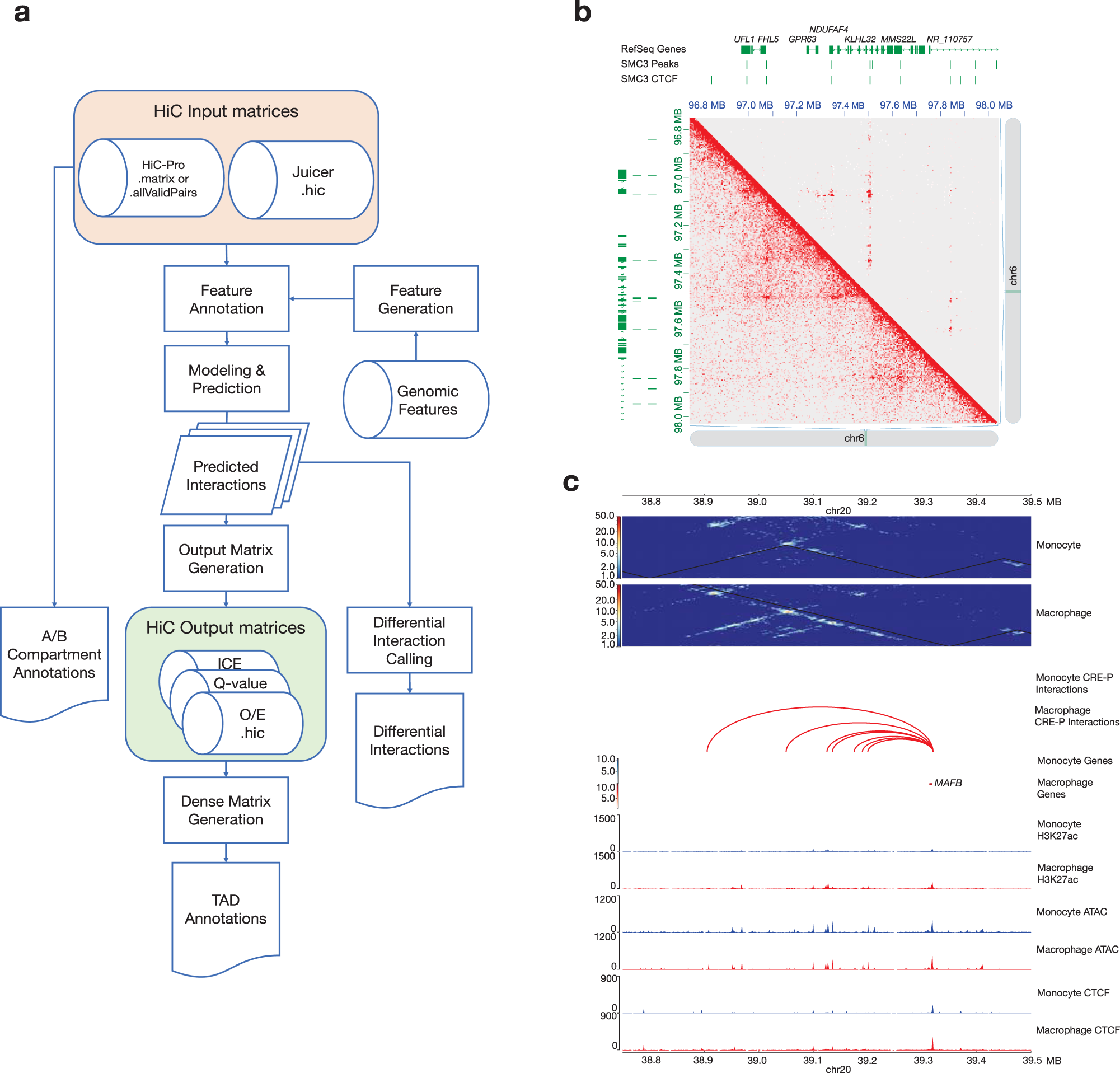
Hic Dc Enables Systematic 3d Interaction Calls And Differential Analysis For Hi C And Hichip Nature Communications

Application Of Hi C And Other Omics Data Analysis In Human Cancer And Cell Differentiation Research Sciencedirect
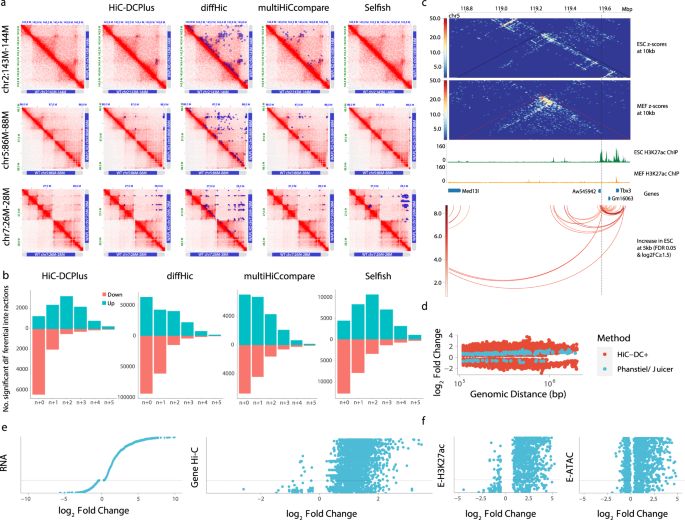
Hic Dc Enables Systematic 3d Interaction Calls And Differential Analysis For Hi C And Hichip Nature Communications

Comparative Hi C Reveals That Ctcf Underlies Evolution Of Chromosomal Domain Architecture Cell Reports
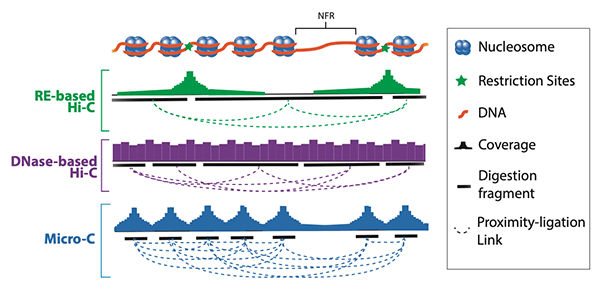
Dovetail Micro C Kit Dovetail Genomics

Chromosome Conformation Capture Wikipedia
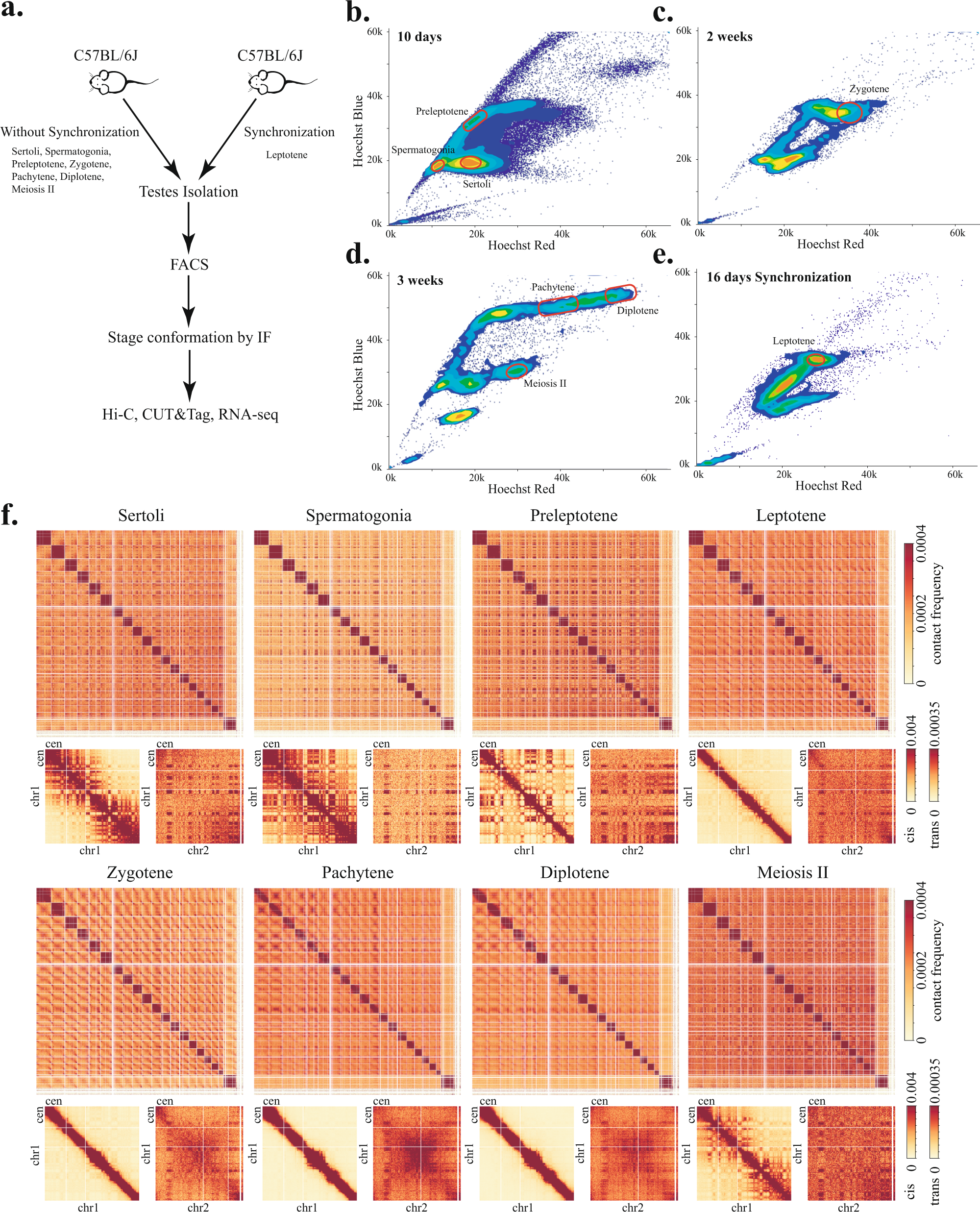
Stage Resolved Hi C Analyses Reveal Meiotic Chromosome Organizational Features Influencing Homolog Alignment Nature Communications


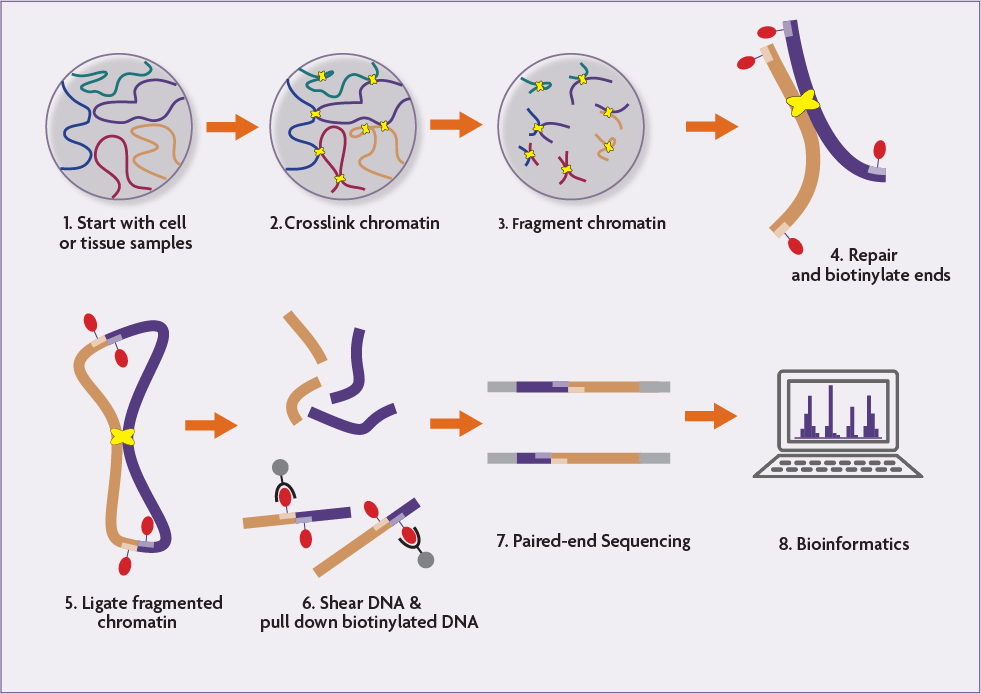
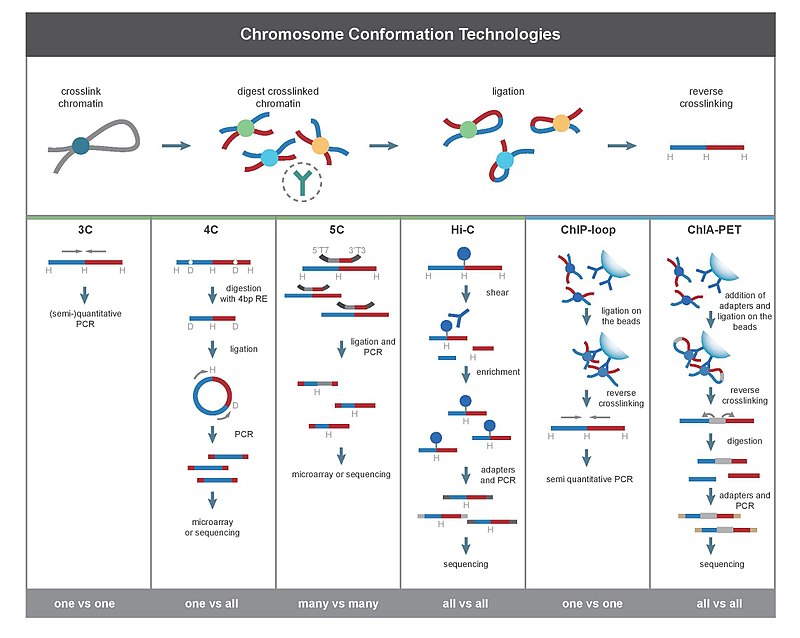
Comments
Post a Comment